News
Exploring the source of AI training data: biopharmaceutical mutation library to help!
"Garbage in, Garbage out", high-quality data has always been considered the basis for AI model training. So what is "high-quality data"? How are these data obtained? Today we would like to introduce an article published in Nature Biomedical Engineering by Sai T Reddy's team from ETH Zurich in 2021: "Optimization of therapeutic antibodies by predicting antigen specificity from antibody sequence via deep learning". The article shows that the biomarker data obtained through the screening of biopharmaceutical mutation libraries is high-quality data that is very suitable for AI training. The authors of the later articles: Derek M. Mason, Simon Friedensohn, and Cédric R. Weber joined deepCDR Biologics, focusing on the application of AI technology in antibody optimization.
Foreword
At present, the multi-parameter optimization of antibodies is very challenging, mainly including expression level, viscosity, pharmacokinetics, solubility and immunogenicity, which also makes us need to spend a lot of time and cost on antibody optimization and screening. In April 2021, Sai T Reddy's team at ETH Zurich published this article, using deep learning to perform high-throughput optimization screening of therapeutic antibodies (full-length IgG) in mammalian cells (Figure 1). Using a mammalian display cell line expressing the therapeutic antibody trastuzumab (Herceptin), the authors combined CRISPR-Cas9-mediated homology-directed repair (HDR) to introduce site-directed repair in the complementarity-determining region 3 (CDRH3) of the variant heavy chain The mutation library is designed by DMS to screen out whether it has binding ability to human epidermal growth factor receptor 2 (HER2). Then use deep learning to predict the specificity of HER2, so as to screen out the optimized drug candidates.
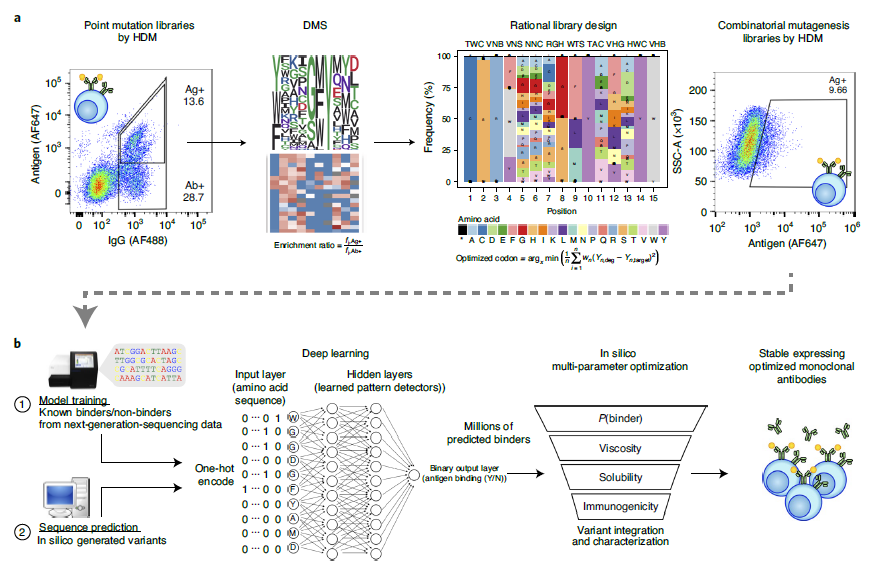
Figure 1. Antibody target specificity prediction using deep learning
Methods and Results
DMS based on CRISPR-Cas9-mediated homology-directed mutagenesis.The amino acid sequence of antibody CDRH3 is a key determinant of antigen specificity, and the authors generated mutation libraries by transfecting gRNA against CDRH3 and a pool of combinatorial templates with NNK degenerate codons contained in single-stranded oligonucleotides (ssODNs) . Then, the population of surface IgG was screened and expressed by FACS, and deep sequencing was performed, and the enrichment rate of amino acids was calculated (Figure 2). CRISPR-Cas9-mediated homology-directed mutagenesis was used to generate repertoires containing CDRH3 variants in mixed tumor cells unable to bind trastuzumab variants of the HER2 antigen.
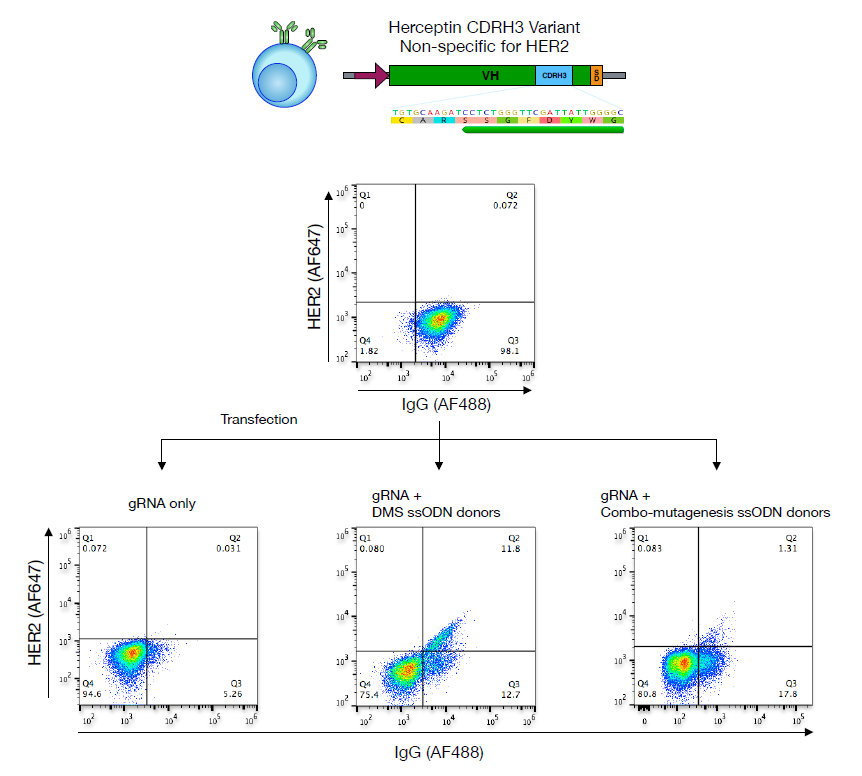
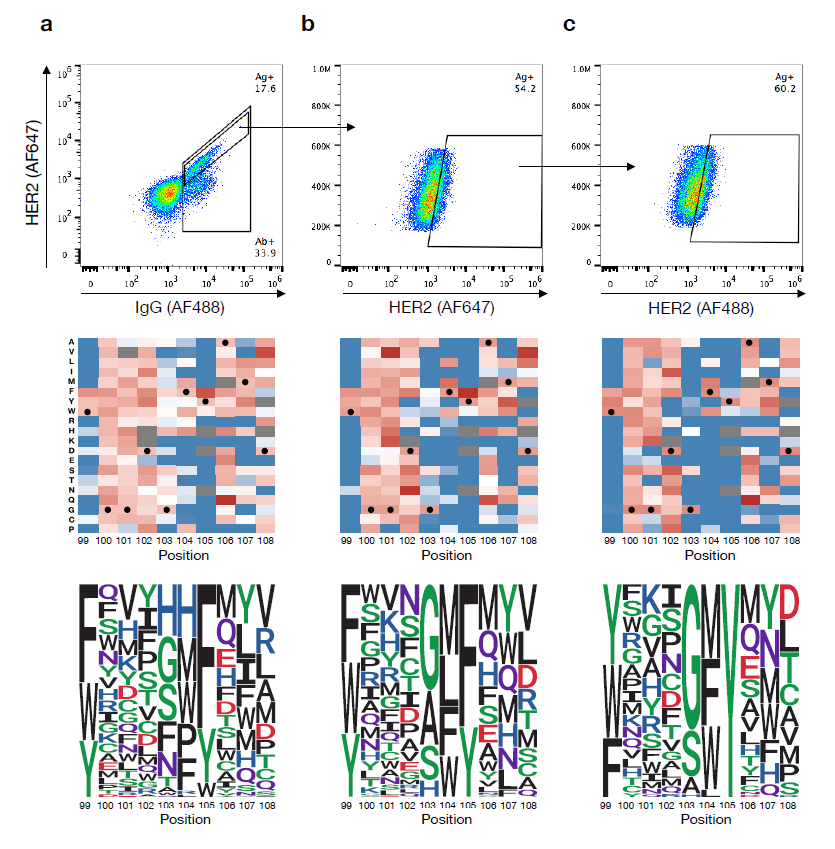
Figure 2. DMS based on CRISPR-Cas9-mediated homology-directed mutagenesis
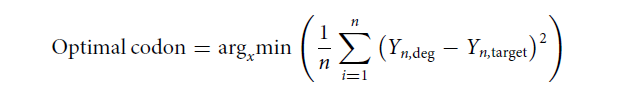
Yn,target is the specific amino acid frequency of a specific site obtained by deep sequencing, and is the frequency of a specific amino acid encoded by a given degenerate codon set at a specific site, and n is the type of amino acid, that is, 20. The theoretical protein sequence space of the combinatorial library is 7.17×10^8, which is far greater than the diversity of the single-site DMS library. The authors isolated antigen-binding cells through two rounds of FACS enrichment, and performed deep sequencing on the antigen-binding and non-antigen-binding cell populations . The sequencing data in the combinatorial library identified 11,300 binders and 27,539 non-binding entities, respectively, and these sequence variations accounted for only 0.0054% of the theoretical protein sequence space of the combinatorial mutant library (Fig. 3).

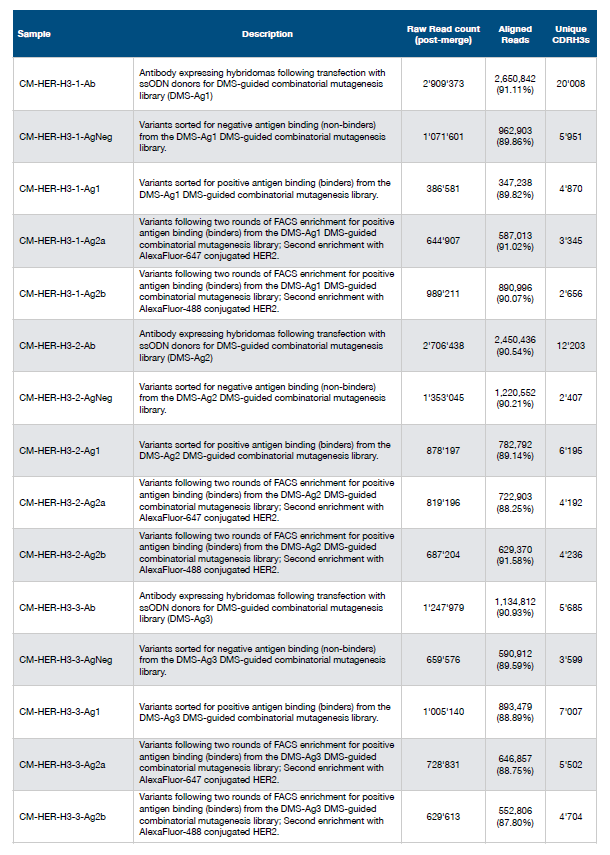
Figure 3. Sequence-based mutation analysis
Sequence-based machine learning and deep learning models for predicting antibody specificity.
The authors set out to develop and train sequence-based machine learning and deep learning models to convert amino acid sequences into input matrices by one-hot encoding after deciphering deep sequencing data of binding and nonbinding CDRH3 variants. Previously, the authors surveyed a series of models to evaluate their accuracy and precision in classifying binders and nonconjugates from existing sequencing data: one-body and two-body logistic regression, k-nearest neighbors, support vector machines (linear and Gaussian kernel), standard artificial neural network, long-short-term memory recurrent neural network (LSTM-RNN) and convolutional neural network (CNN), and found that the CNN deep learning model is superior to other test models, and can accurately predict unseen test data Classify (Figure 4).
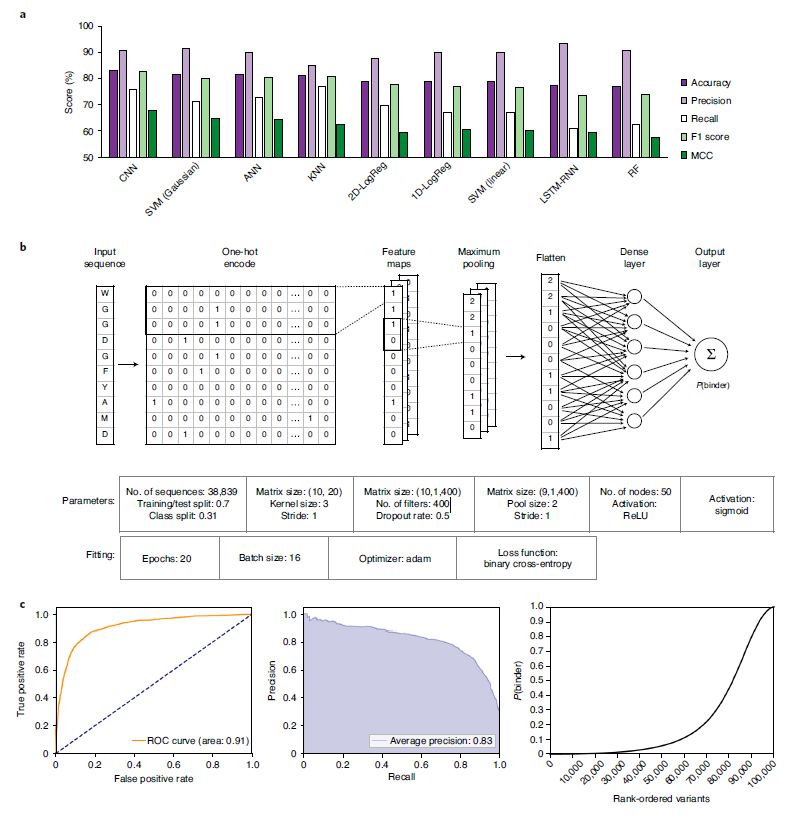
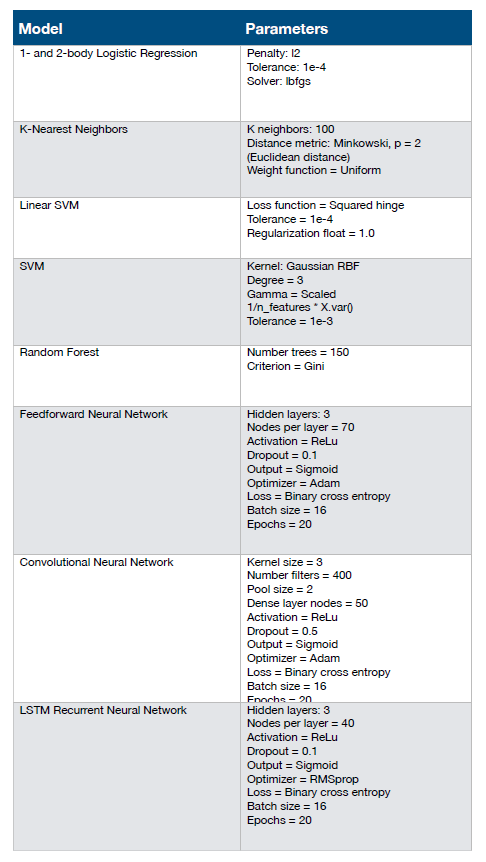
Figure 4. The deep learning model accurately predicts antigen specificity
The authors assessed the false positive and negative rates of the model by detecting the binding of randomly selected sequences to the target antigen by BLI. It was found that the binding sequences all maintained a remarkably high affinity to the HER2 antigen. However, non-binding data for three of the nine sequences also maintained affinity for the HER2 antigen (Fig. 5), suggesting an inaccuracy in the data set that could be exploited in the future by using additional sorting strategy to solve this problem.
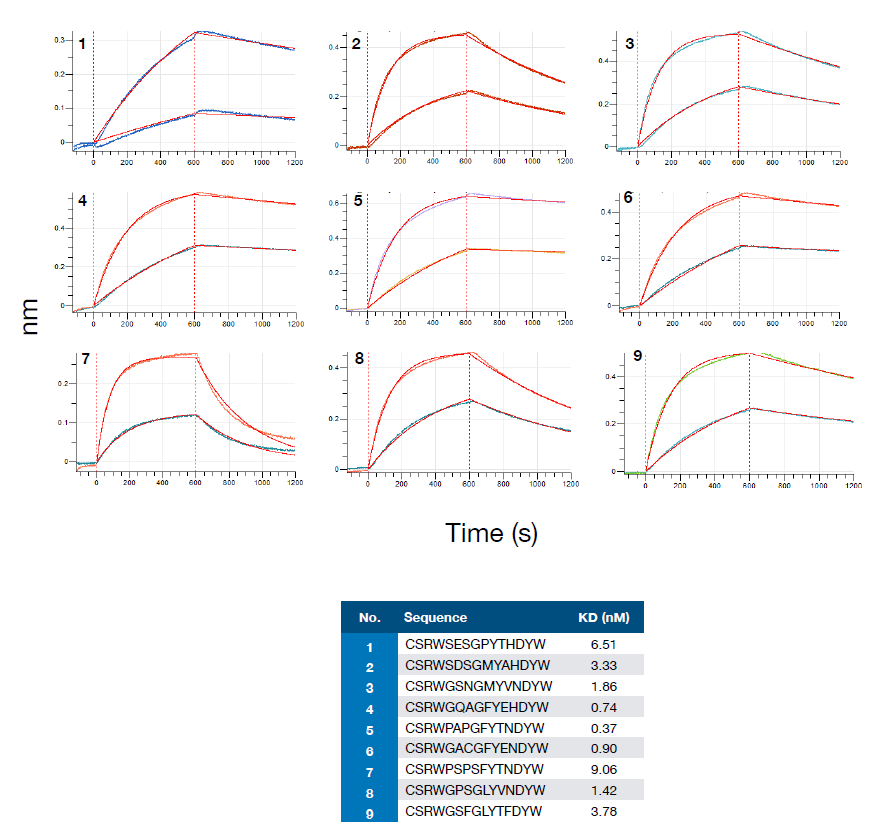

Figure 5. Randomly selected sequences from the experimental dataset for BLI analysis
In the final step of model validation, the authors trained a neural network that showed indiscriminate sequence classification on unseen test data (Figure 6), suggesting that a network trained on correctly classified data can recognize learned patterns.
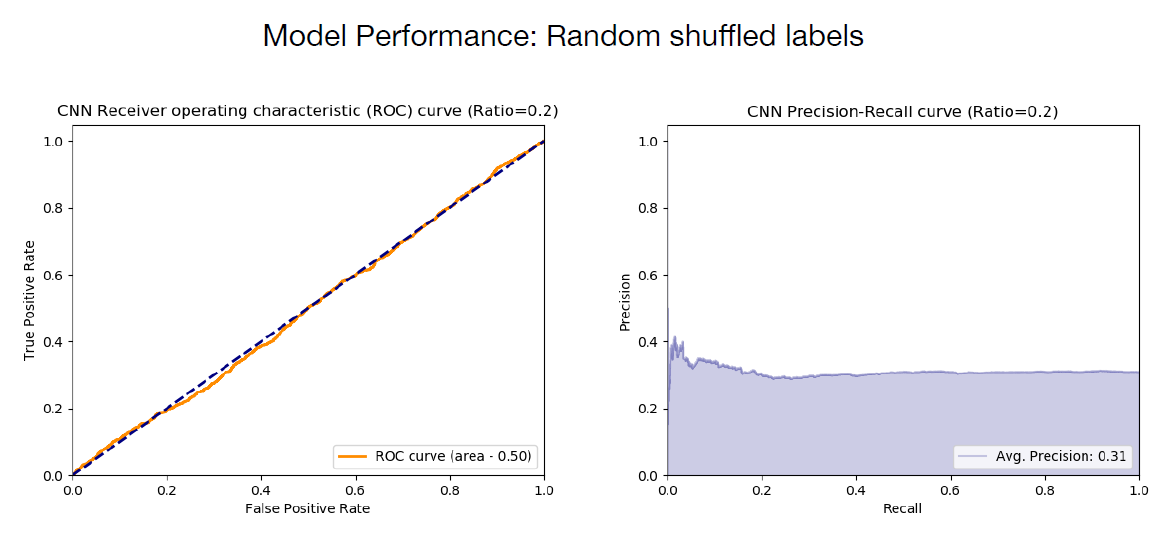
Figure 6. Model performance
Perform experimental characterization of the selected sequences and screen out the best candidate sequences.
The authors used CRISPR-Cas9 HDR technology to generate a stable cell bank, performed single-cell sorting, and further characterized the monoclonal variants, and finally identified 55 mutants. Their expression levels were then measured from cell supernatants using BLI and showed varying degrees of antibody titers, with five variants showing comparable or better expression than trastuzumab. After purification, thermal stability was tested by fluorescence measurement, and all 10 variants were found to be comparable or better in thermal stability than trastuzumab (Figure 7). The authors selected a 15-amino acid fragment of each variant and wild-type trastuzumab based on regions predicted to have higher immunogenic potential using the NetMHCII prediction method and found that variants 1 and 3 showed no significant T cell activation, showed a reduction in its immunogenicity. Moreover, the expression level of the variant 1 sequence is equivalent to that of trastuzumab, and the thermal stability is higher. Compared with the original sequence of trastuzumab, this variant shows obvious risk-removing immunogenicity potential.
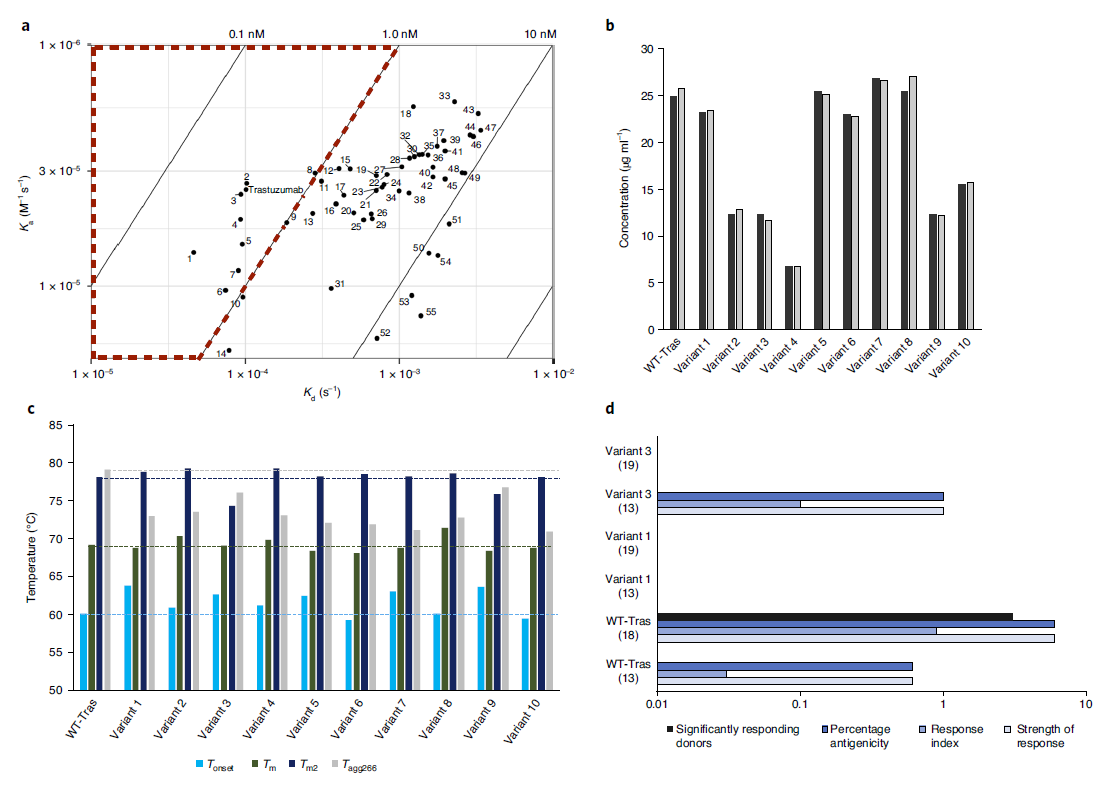

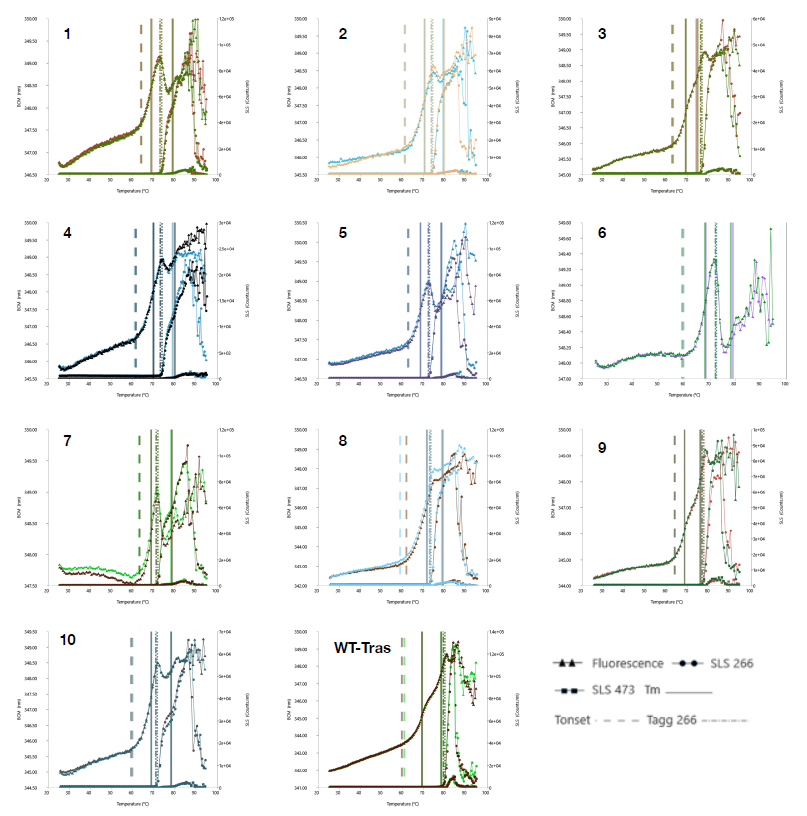
Figure 7. Screening out the best candidate sequences
In Conclusion
In this paper, the author obtained the HER2-specific antibody sequence through deep learning and antigen-specific detection, combined with multiple bioinformatics methods for target screening, and obtained a highly optimized HER2 leader sequence. This paper introduces in detail the method of generating high-quality antibody libraries by optimizing gene editing technology. The developed method based on deep learning can identify highly specific antigenic sequences, which greatly saves time and cost, and greatly reduces the risk of downstream clinical development. , this research is of great significance in promoting antibody engineering and drug development, and is expected to provide new methods and strategies for the development of precision medicine and personalized treatment.
Liu Bo said:
Every AI company is looking for high-quality data!
What exactly is high-quality data? In this article, scientists from ETH Zurich gave their definitions: labeled (antigen specificity in the article), large data volume (about 104 variants in the article), obtained by real screening (article Antibody amino acid sequence data screened and enriched from Combinatorial mutagenesis libraries. Using these high-quality data, the author trained and evaluated various machine or deep learning models, and finally confirmed the performance of the convolutional neural network (CNN) on multiple indicators such as accuracy and precision. Most excellent, suitable for antibody optimization process based on amino acid sequence.
How can we obtain high-quality data? Deep learning (such as CNN in the article) has high requirements on the label and quantity of data. However, the data obtained from the public database has a relatively single label; while the data obtained from the patent literature is fragmented and the labels are out of order; It is not convenient, and there are many limitations in the training of AI algorithm models.
This article uses a method of obtaining high-quality data using antibody mutation library technology: firstly, through random mutation or rational design mutation, the antibody mutation library is displayed on the surface of mammalian cells, and the specific binding to the antigen is enriched by flow sorting antibody amino acid sequence. Usually, the antibody sequence obtained from the antibody mutation library will have the following characteristics:
1) Sequences are labeled. This article is based on the screening of antigen binding ability, so the tag carried by the sequence is "antigen specific". If we change the screening conditions, such as screening based on expression level, based on functional screening, or based on stability screening, then the obtained sequence will carry "expression level", "activation/blocking ability", "structural stability", etc. different labels.
2) The sequence is continuous. By performing saturation mutation or design mutation on a single site or multiple sites of an antibody, continuous amino acid changes at a certain site or multiple sites can be obtained, helping AI to make continuous and subtle changes at these sites to study.
3) Sequences are sorted. If we are screening the affinity of antibodies, after performing NGS sequencing on the enriched sequences, we can obtain the number and frequency of each antibody sequence. In theory, the larger the number and the higher the frequency, the higher the degree of enrichment. Higher, the higher the affinity with the antigen, so that the sequences obtained by NGS (usually tens of thousands to hundreds of thousands of sequences, depending on the depth of sequencing) can be relatively accurately sorted.
4) Sequences can be grouped into positive/negative data, such as the amino acid sequences of binder and non-binder antibodies in the article.
Therefore, compared with public databases and literature data, the data generated by artificial mutation library screening has better accuracy and continuity, which can also be understood as higher "resolution".
Although the article only carried out the work of AI antibody optimization in the label or dimension of antigen specificity, it proved the feasibility and effectiveness of "using label data produced by artificial mutation library" for AI training. The CRISPR-Cas9 mutation and mammalian cell display in this article are just one way to construct an artificial mutation library. If it is extended to more mutation technologies and display technologies, such as fully synthetic library technology and phage display technology, the number of sequences generated and the sequence size There will be more types of labels, such as various labels such as high temperature resistance, pH sensitivity, and protease resistance. These sequence data will also be more helpful for AI learning and training.
Liu Jianghai
Ph.D., University of Saskatchewan, Canada
The founder and CEO of Shengshi Junlian Company, once studied as a postdoctoral fellow at the Therapeutic Antibody Resource Center in Saskatchewan, Canada. Successively won the Sichuan Province "Thousand Talents Program (Entrepreneurship Leader), "Rongpiao Talents" Program, and "Golden Panda Talents" Program. Has rich practical experience in the fields of total synthetic library technology, biopharmaceutical library design and construction, antibody discovery and optimization With experience, he has led the preclinical development of multiple monoclonal antibodies, double antibodies, and CAR-T drugs.
