News
Shengshi Junlian participated in the development of Zhang Kang's team's new artificial intelligence PPI prediction model to improve the research and development capabilities of macromolecular drugs
Protein-protein interaction (PPI) is the process of combining two or more proteins and participates in the regulation of many aspects of life processes. How to establish an efficient and accurate PPI prediction model has always been a hot and difficult point in AI research. Professor Zhang Kang, the AI science consultant of Chengdu Shengshi Junlian Biotechnology Co., Ltd., recently published "Deep-learning-enabled protein–protein interaction analysis for prediction of SARS-CoV-2 infectivity and variant evolution" in "NATURE MEDICINE". This paper describes an innovative artificial intelligence PPI prediction model - "UniBind". UniBind integrates and analyzes data from various experimental sources and modalities, and based on the rules of biological evolution, it predicts possible emerging new coronavirus mutant strains. More importantly, for multiple ACE2 mutants predicted by UniBind, wet experiments confirmed that the binding force to RBD was 1000 times higher than that of the wild type. ABLINK BIOTECH participated in the protein molecular design and wet experiment verification in the article.
Article overview
The rapid development of deep mutational scanning techniques in recent years has generated large-scale datasets that have the potential to reveal intrinsic features of proteins. Combined with other large-scale proteomics and biochemical databases, such as SKEMPI 2.0, the large amount of data provides the possibility for AI modeling and PPI prediction.
UniBind is a multi-task learning method. By integrating protein structure, affinity data and heterogeneous biological information, it can effectively predict the binding affinity of SARS-CoV-2 mutations on human and other species ACE2 receptors and antibodies. After systematic testing and experimental verification on multiple benchmark datasets, UniBind can accurately and scalablely predict the impact of SARS-CoV-2 mutations on the binding affinity of human ACE2 receptors, the impact on neutralizing antibodies, and predict the virus Future variation trends.
UniBind features multi-task learning and model ensemble training modes to address data heterogeneity. Taking the prediction of 1-4 point mutations of SARS-CoV-2 RBD as an example, multi-point prediction can generate more constraints on PPI, thereby more effectively predicting future virus evolution and immune escape. Likewise, composite scores for antibody affinity and immune escape against known variants were consistent with clinical observations.
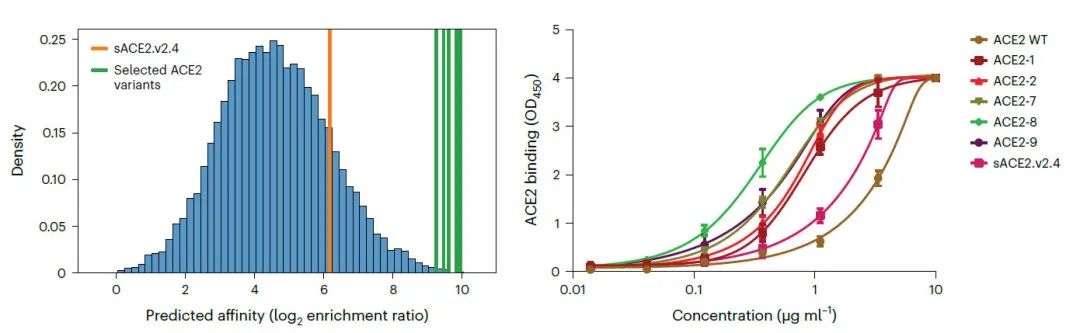
This model can be extended to predict the effect of ACE2 mutations on virus affinity. Based on an evolutionary approach based on computational simulations, UniBind generated 13,913 ACE2 variants with higher affinity than wild-type. Five ACE2 variants with high model prediction scores were selected for wet assay validation and showed 100-1000-fold improvement in binding affinity to the S protein (bottom panel), these results highlight the accuracy of UniBind in PPI prediction .
In summary, UniBind provides a general framework for predicting protein interactions, which is very suitable for the development of macromolecular drugs in the field of biomedicine.
Professional Review
Xin Zeng (Chief Technology Officer):
PPI can be described as the next killer breakthrough in protein AI prediction in the "post-AlphaFold2 era". This paper chooses the subdivision of COVID-19, which is a rapidly growing data field, as the entry point. With the researchers' excellent understanding of biology and informatics, biological data from multiple sources are integrated and used. It can be said that limited data is used to improve data quality. A typical case of further improving the capabilities of AI models. In addition, wet experiments are still the gold standard for testing dry experiments. For the AI+ application field, which is chasing hot spots and hot spots change every few months, the engineering ability of dry and wet combined with a virtuous cycle is also indispensable.
Chen Huang (Chief Information Officer):
AI For Biotechical has made major breakthroughs in the past few years. With the continuous application of AI to the research of PPI, on the one hand, it will rapidly iterate on the AI model, and on the other hand, the progress of biotechnology, such as the establishment of a trillion-level diverse phage library Combined with NGS sequencing, the growth rate of biological data will also increase exponentially. It is expected that AI For Bio will fundamentally subvert drug design in the next 2-3 years. At the current stage, AI models and biological experiment methods complement each other. AI models provide predictions that greatly reduce time costs and improve efficiency. Biological experiment methods verify conclusions and clarify data labels. This paper establishes the UniBind model, uses experimental methods to verify the conclusions of the model, and makes scientific predictions on the infection and evolution of the new coronavirus. The biggest highlight is the processing of massive biological heterogeneous data. The paper recognizes that although modeling technology It is very important, but it is the data that really determines the quality of the model. The model in this paper is established based on the data related to the specific problem, which realizes the closed-loop verification of the experimental data and the AI model. In addition, if there are high-quality massive antibody label data, and then establish a hyperparameter adaptation model for multi-task and heterogeneous integration, UniBind can also develop into a general model for antibody drug design.
Character Introduction
Zhang Kang
AI scientific consultant of Shengshi Junlian Company, tenured full professor of ophthalmology and human genetics at the University of California, San Diego, and the first dual-degree Ph.D. in medicine and genetics from China at Harvard University and Massachusetts Institute of Technology. Selected into the national "Thousand Talents Program". He is currently a professor of the Faculty of Medicine of the Macau University of Science and Technology, an ophthalmologist at the University Hospital of Macau, and the deputy dean of the Faculty of Medicine of the Macau University of Science and Technology. Based on large-scale clinical data analysis and deep machine learning, the research team led by him has published many breakthrough articles on the application of AI technology in top journals such as CELL, Nature, and Nature Medicine.
Zeng Xin
Chief Technology Officer of Shengshi Junlian Company. Ph.D. in Structural Biology from Tsinghua University. He has been selected into the "Rongpiao Talents" Program of Chengdu City and the "Golden Panda Talents" Program of High-tech Zone. He once published protein structure research work in Nature as the co-first author, and has rich first-line experience in the fields of structure-based biopharmaceutical development and AI Workflow design.
Huang Chen
Chief Information Officer of Shengshi Junlian Company. Master of Biomechanics from Sichuan University, 10+ years working experience in Microsoft and Oracle, core member of "Rong Piao Top Team". Has extensive experience in data management, integration, and fusion.If you are interested in understanding the original text, please contact us~
